

| Statistics Toolbox |   |
Create hierarchical cluster tree.
Syntax
Z = linkage(Y) Z = linkage(Y,'method')
Description
Z = linkage(Y)
Y, is the distance vector output by the pdist function, a vector of length 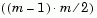 -by-1, where m is the number of objects in the original dataset.
-by-1, where m is the number of objects in the original dataset.
Z = linkage(Y,'method')
'method', where 'method' can be any of the following character strings that identify ways to create the cluster hierarchy. Their definitions are explained in Mathematical Definitions.
The output, Z, is an (m-1)-by-3 matrix containing cluster tree information. The leaf nodes in the cluster hierarchy are the objects in the original dataset, numbered from 1 to m. They are the singleton clusters from which all higher clusters are built. Each newly formed cluster, corresponding to row i in Z, is assigned the index m+i, where m is the total number of initial leaves.
Columns 1 and 2, Z(i,1:2), contain the indices of the objects that were linked in pairs to form a new cluster. This new cluster is assigned the index value m+i. There are m-1 higher clusters that correspond to the interior nodes of the hierarchical cluster tree.
Column 3, Z(i,3), contains the corresponding linkage distances between the objects paired in the clusters at each row i.
For example, consider a case with 30 initial nodes. If the tenth cluster formed by the linkage function combines object 5 and object 7 and their distance is 1.5, then row 10 of Z will contain the values (5, 7, 1.5). This newly formed cluster will have the index 10+30=40. If cluster 40 shows up in a later row, that means this newly formed cluster is being combined again into some bigger cluster.
Mathematical Definitions
The 'method' argument is a character string that specifies the algorithm used to generate the hierarchical cluster tree information. These linkage algorithms are based on various measurements of proximity between two groups of objects. If nr is the number of objects in cluster r and ns is the number of objects in cluster s, and xri is the ith object in cluster r, the definitions of these various measurements are as follows:
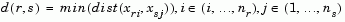
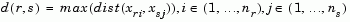
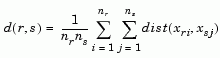
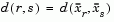
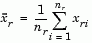
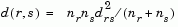
where
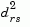 is the distance between cluster r and cluster s defined in the Centroid linkage. The within-group sum of squares of a cluster is defined as the sum of the squares of the distance between all objects in the cluster and the centroid of the cluster.
is the distance between cluster r and cluster s defined in the Centroid linkage. The within-group sum of squares of a cluster is defined as the sum of the squares of the distance between all objects in the cluster and the centroid of the cluster.
Example
X = [3 1.7; 1 1; 2 3; 2 2.5; 1.2 1; 1.1 1.5; 3 1]; Y = pdist(x); Z = linkage(y) Z = 2.0000 5.0000 0.2000 3.0000 4.0000 0.5000 8.0000 6.0000 0.5099 1.0000 7.0000 0.7000 11.0000 9.0000 1.2806 12.0000 10.0000 1.3454
See Also
cluster, clusterdata, cophenet, dendrogram, inconsistent, pdist, squareform
 | lillietest | logncdf |  |